folding@home
PCLinuxOS Team # 50619
What exactly is it? It is a scientific research project conducted by Stanford University. Instead of renting time on a supercomputer to run computations, they seek individuals who will volunteer their computer's idle time to the research project. A good description of what "folding" is at:
You can help scientists studying these diseases by simply running a piece of software.
Folding@home is a distributed computing project -- people from throughout the world download and run software to band together to make one of the largest supercomputers in the world. Every computer takes the project closer to our goals. Folding@home uses novel computational methods coupled to distributed computing, to simulate problems millions of times more challenging than previously achieved.
Protein folding is linked to disease, such as Alzheimer's, ALS, Huntington's, Parkinson's disease, and many Cancers. Moreover, when proteins do not fold correctly (i.e. "misfold"), there can be serious consequences, including many well known diseases, such as Alzheimer's, Mad Cow (BSE), CJD, ALS, Huntington's, Parkinson's disease, and many Cancers and cancer-related syndromes.
What is protein folding?
Proteins are biology's workhorses -- its "nanomachines." Before proteins can carry out these important functions, they assemble themselves, or "fold." The process of protein folding, while critical and fundamental to virtually all of biology, in many ways remains a mystery."
The link to download the client is at:
http://www.stanford.edu/ … /FAH6.02-Linux.tgz.
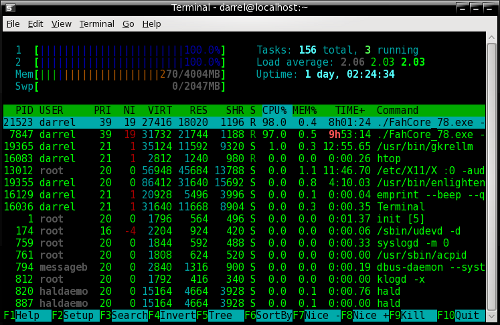
The link is to the 32-bit client, since PCLinuxOS does not yet have a 64-bit OS. If your computer has multiple cores, you can run one client per core. Each client will have a different machine ID number, in order the keep the running tasks separate. Each client will be run from within its own folder. There is no GUI for running the client. It is run in a terminal, or as a service. Because I never know what project I will be doing on any of my computers, I run the program in a quake-like drop-down terminal, rather than as a service. The possible choices from our repositories are guake, tilda, and yakuake. The screenshot below is of a client running in one pane of the guake terminal.
After downloading the client, open any terminal and do the following: (Note: I am using fold1 as an example folder name.)
cd fold1 ./fah6 -configonly
This will start the client's configuration runtime options. You can override the configuration at any time by starting folding with a command line option. For example,
./fah6 -advmethods
One of the configuration options is adding a personal passkey. What is a passkey?
Quoted from: http://folding.stanford.edu/English/FAQ-passkey.
"The passkey, a new feature beginning with the v6.0 FAH client, is a unique identifier that ties your contributions directly to you (not just those with your username). The use of a passkey prevents others from cheating using your name. Obtain a passkey from our web site (see below), enter it when you configure the client, and the client and servers will do the rest. You should keep your passkey secret."
To get your passkey, go to:
http://fah-web.stanford.edu/cgi-bin/getpasskey.py.
After configuring the client, start it with ./fah6.
All my clients run 24/7 on desktop machines. The program is so unobtrusive I barely notice it unless I bring the drop-down terminal into focus, run top or htop, or glance at the Gkrellm panel on my desktop. Cruising the web, viewing videos, opening or editing OpenOffice documents is just about as quick with the folding clients running as it would be without them running. The only other time I notice the running clients is if I am downloading something from the web at the same time that a client finishes a work unit. The client will upload the finished results to a Stanford University server with a higher priority than any download process.
Will you join the PCLinuxOS folding team today?